This post is strictly about binary matrices. For matrices of any data type, have a look at this post.
The hamming distance is perhaps the most frequently used distance measure for binary vectors. It is defined on two vectors of equal length as the number of corresponding elements that differ between the two vectors. A special case of the Hamming distance, is the Hamming distance between two binary vectors, which is often used e.g. in telecommunication to count the number of bit flips in a signal, and in bioinformatics as a similarity measure for hierarchical clustering. While this binary Hamming distance calculation between only two vectors is simple and computationally cheap, calculating all pairwise distances between the rows of a matrix quickly becomes prohibitive, as the computation time scales quadratically with the number of rows in a matrix. The following R function represents a basic implementation of calculating the Hamming distance between all rows of a matrix, using a nested for-loop:
hamming_loop <- function(X) {
d <- matrix(0, nrow = nrow(X), ncol = nrow(X))
for ( i in 1:(nrow(X) - 1) ) {
for ( j in (i + 1):nrow(X) ) {
d[i,j] <- d[j,i] <- sum(X[i,] != X[j,])
}
}
d
}
However, in R, the Hamming distance between the rows of a binary matrix can be computed much faster, by exploiting the fact that the dot product of two binary vectors x and (1 – y) counts the corresponding elements where x has a 1 and y has a 0:
hamming <- function(X) {
D <- (1 - X) %*% t(X)
D + t(D)
}
To demonstrate how much faster the matrix multiplication function is than the nested for-loop function, I ran some simulations. In addition to the above two functions, I included the function hamming.distance from the R-package e1071:
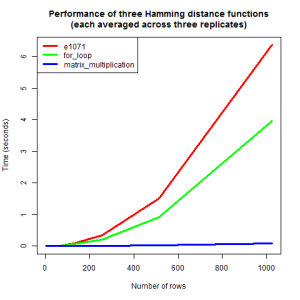
The relatively naive nested for-loop approach is faster than the hamming.distance function from the e1071 package. This is surprising, since the e1071 hamming.distance function is also implemented as a nested loop. Strikingly, the matrix multiplication-based function is by far the fastest function for computing the Hamming distance, the reason being that it does not use any expensive looping at R-level, only efficient lower level linear algebra routines. Note that the above approach is restricted to binary matrices. However, quite conveniently, matrix multiplication can also be used for computing the Hamming distance for matrices with more than two possible values, and other types of elements (character, numeric, …), as explained in this post.
Leave a comment